2019 Volume 2 Issue 2
|
|
INEOS OPEN, 2019, 2 (2), 50–54 Journal of Nesmeyanov Institute of Organoelement Compounds DOI: 10.32931/io1909a |
|
Atomic Force Microscopy of Peptide Nanoparticles Produced by
Tryptic Hydrolysis of β-Casein
Nesmeyanov Institute of Organoelement Compounds, Russian Academy of Sciences, ul. Vavilova 28, Moscow, 119991 Russia
Corresponding author: M. M. Vorob'ev, e-mail: mmvor@ineos.ac.ru
Received 21 February 2019; accepted 12 April 2019
Abstract
Peptide nanoparticles are obtained by the controlled hydrolysis of β-casein by trypsin. The atomic force microscopy studies of the peptide nanoparticles dried on the surface of mica allowed us to estimate their sizes, density, and structural heterogeneity. These nanoparticles appeared to be approximately two times denser than the initial casein micelles. The results obtained are important for the design of new biocompatible and nontoxic macromolecular systems for transporting biologically active compounds.
Key words: peptide nanoparticles, atomic force microscopy, beta-casein, trypsin.
Introduction
Most of biologically active compounds are poorly soluble in water, and they are transported in vivo as complexes with macromolecules, nanoscale associates, and micelles [1]. Solubilization in micelles is one of the promising methods for the targeted delivery of hydrophobic drugs, which improves the efficiency of these compounds [2, 3]. Biocompatible nontoxic protein micelles can significantly increase the solubility of hydrophobic compounds. This is used to develop transport systems, in particular, on the basis of relatively cheap milk proteins, including amphiphilic β-casein (β-CN) [4].
In contrast to globular proteins, casein proteins (αs1-, αs2-, β- and κ-casein) do not have fixed three-dimensional tertiary structures; they have only secondary structures which predetermine a set of preferred conformational states of polypeptide chains. The structures and properties of casein proteins and their aggregates (micelles) in aqueous media are defined by a rheomorphic model [5]. According to this model, casein proteins are distributed over those conformational states making them highly hydrated. In turn, casein micelles are loose, so that the content of water in these micelles composes 2–3 g per 1 g of protein [6].
Earlier we have studied the kinetics of hydrolysis (proteolysis) of β-casein by trypsin and the formation of nanoparticles which consisted of the products of enzymatic hydrolysis [7, 8]. This process was firstly demonstrated by static light scattering using the dissymmetry method (Debye method) for continuous recording of the nanoparticle concentrations and sizes in the range of 10–200 nm during proteolysis [8]. For this purpose, light scattering intensities I(45o) and I(135o) were measured directly in the reaction mixture for 2–3 h with the time resolution of 1–2 min [8, 9]. Since nanoparticles are firstly formed during proteolysis and then are degraded under action of the same enzyme, it is important to find the time interval when the maximum number of nanoparticles is formed. It was shown that the peptide nanoparticles obtained differ from the original casein micelles both in sizes and in the mode of interaction with low-molecular hydrophobic compounds [10]. However, to date there has been no detailed study of these nanoparticles by atomic force microscopy (AFM).
AFM working principle is based on the interaction of a sharp needle with particles lying on the surface [11]. An important advantage of AFM is the possibility to study the shape, size and structural heterogeneity of particles at the molecular level [12]. In the microscopic studies of polymeric and biological objects, it is preferred to avoid additional staining, metal shading, or freezing of the samples for achieving high resolution. Often the images must be taken under ambient conditions, which is possible with AFM technique. AFM has been widely used to characterize the morphology of nanostructures formed by milk proteins [13].
The goal of the present work was to investigate the peptide nanoparticles obtained by controlled tryptic hydrolysis of β-casein using AFM. The method in use implied termination of hydrolysis with a soybean trypsin inhibitor and application of a simple technique for preparing a sample on the mica surface via air drying. This sample preparation technique allows one to achieve the highest resolution in AFM studies [14].
Results and discussion
The peptide nanoparticles were obtained by limited hydrolysis of peptide bonds in β-CN (disordered oligopeptide bearing 209 amino acid residues) by trypsin (EC 3.4.21.4) in 50 mM phosphate buffer. This serine protease provides hydrolysis of some peptide bonds Arg-X and Lys-X, wherein the amount of hydrolyzed bonds (degree of hydrolysis) may be varied by changing the hydrolysis time. The peptide nanoparticles, which differ from the original casein micelles, were obtained during the hydrolysis for 5–65 min, depending on the concentrations of the protein and proteolytic enzyme. The analysis of the resulting nanoparticles was performed by AFM using a FemtoScan microscope. All the samples taken from the reaction mixture were diluted 10 times, applied to the freshly cleaved mica surface and dried in air (all the samples were prepared under the same conditions). A visual comparison of the topographic images of the original micelles and the resulting nanoparticles reveals significant differences: the original micelles form a chaotic network (Fig. 1), while the modified ones represent separate axisymmetric particles with clearly defined edges and smooth surface (Fig. 2).
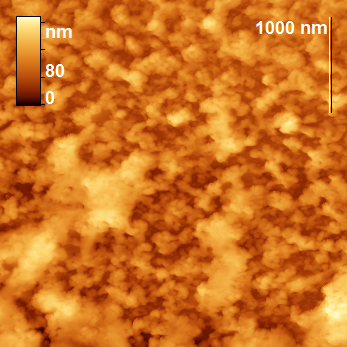
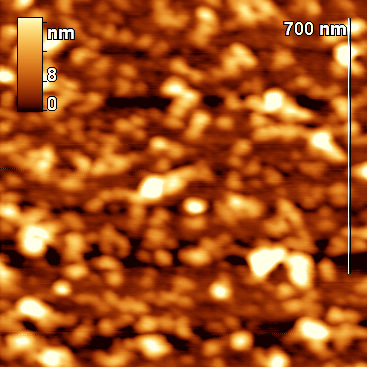
Figure 1. Topographic images of the original particles (β-casein micelles).
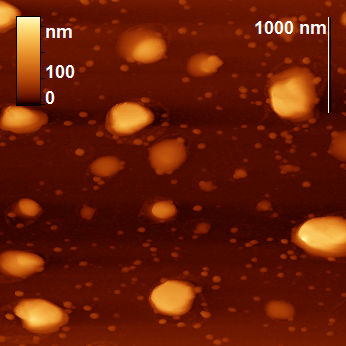
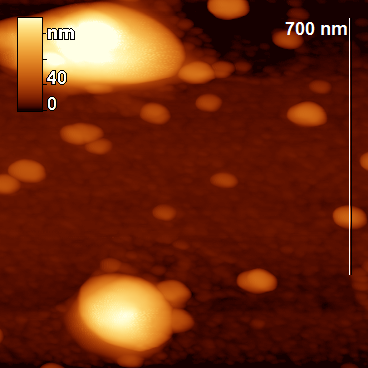
Figure 2. Topographic images of the particles obtained by proteolysis.
To study the physicochemical properties of peptide nanoparticles, it is necessary to terminate proteolysis (inhibit the enzyme) to stabilize the particle size distribution, since further proteolysis leads to degradation of the particles. A comparison of the termination of hydrolysis was carried out using a low-molecular inhibitor, phenylmethylsulfonyl fluoride (PMSF), and a high-molecular inhibitor, a soybean inhibitor of trypsin. The use of the soybean inhibitor (the mass ratio of the inhibitor to trypsin was 3:1) ensured that the hydrolysis was stopped and the particle distribution was unchanged according to AFM. The use of PMSF led to the redistribution of nanoparticles in size according to AFM. Therefore, we decided to carry out a detailed analysis of the surface and particle shape with the termination of proteolysis using the soybean inhibitor.
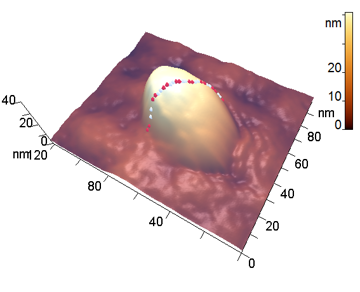
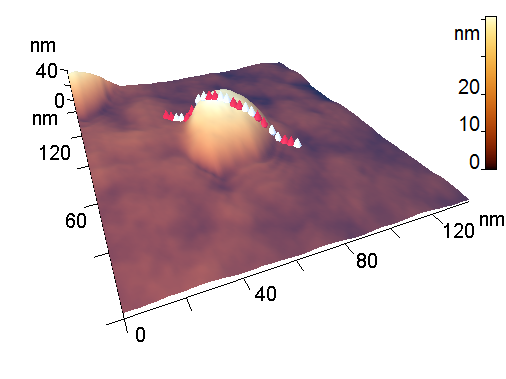
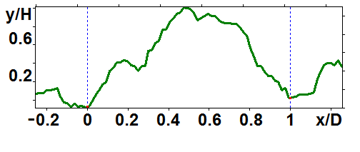
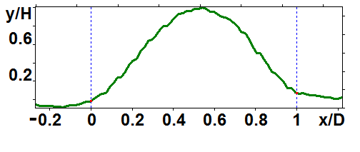
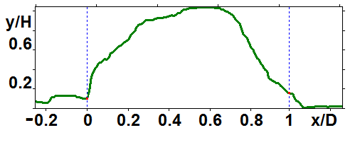
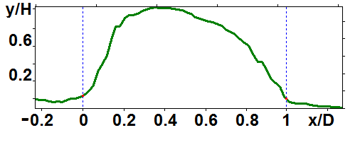
The analyzed nanoparticles were categorized into the large nanoparticles with the diameter (D) of several hundred nm and the medium ones with the diameter of up to 100 nm. The analysis of the smaller particles (20–30 nm) required the use of another procedure for the dilution of the reaction mixture for sample preparation and is not considered in this paper. The profiles of the randomly chosen particles were analyzed by scanning along the lines indicated in Fig. 3a,b (large particles) and Fig. 4a,b (medium particles). The particle profiles in dimensionless coordinates x/D and y/H, where D is the diameter and H is the maximal height, turned out to differ for the initial micelles and the nanoparticles obtained by hydrolysis. For the initial micelles, the profiles have the forms (Fig. 5a,b) that differ from those of the modified ones (Fig. 6a,b). The latter profiles are more likely to be semicircular both for the large nanoparticles (Fig. 6a) and for the medium ones (Fig. 6b).
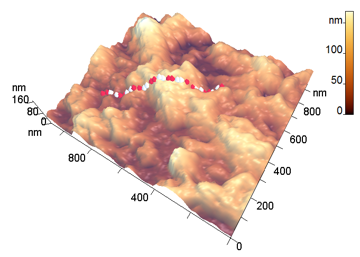
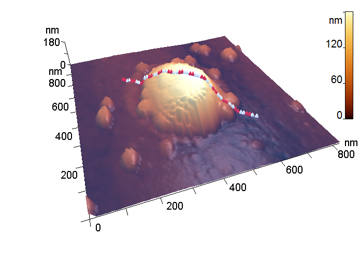
a b
Figure 3. 3D-Images of the original (a) and large peptide particles obtained by proteolysis (b).
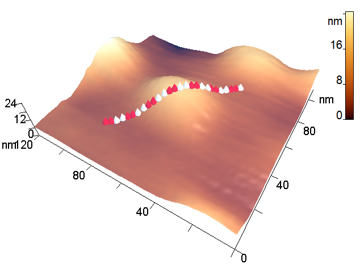
a b
Figure 4. 3D-Images of the original (a) and medium peptide particles obtained by proteolysis (b).
a b
Figure 5. Profiles of the original particles: large (a) and medium (b) ones.
a b
Figure 6. Profiles of the peptide nanoparticles modified by proteolysis: large (a) and medium (b) ones.
An essential difference between the original and modified particles was detected in their densities, which were evaluated as the ratio of the heights of the dried particles to their diameters H/D (Table 1). For the modified particles, this parameter composed 0.36 ± 0.12 (large nanoparticles) and 0.67 ± 0.15 (medium nanoparticles), and for the initial micelles 0.25 ± 0.10 (large nanoparticles) and 0.27 ± 0.06 (medium nanoparticles). Thus, the modified particles were found to be denser, especially the medium ones which were approximately two times denser. These changes in H/D parameter for the dried nanoparticles can be illustrated by a simple scheme assuming that the hydrolysis leads to shrinkage of micelles in solution (Fig. 7). In fact, the proteolytic modification of casein micelles may include theincorporation of hydrophobic peptides, the products of hydrolysis of the peptide chain. This process may also increase the micelle density.
Table 1. Comparison of the original β-CN micelles with the nanoparticles obtained by tryptic hydrolysis
|
Sample |
Particle size |
D (nm) |
H (nm) |
H/D |
|
Initial β-CN micelles |
Large |
460 |
110 |
0.24 |
|
370 |
150 |
0.41 |
||
|
605 |
130 |
0.21 |
||
|
340 |
90 |
0.26 |
||
|
430 |
65 |
0.15 |
||
|
|
|
0.25±0.10 |
||
|
Medium |
49 |
10 |
0.20 |
|
|
43 |
12 |
0.28 |
||
|
67 |
23 |
0.34 |
||
|
48 |
11 |
0.23 |
||
|
67 |
21 |
0.31 |
||
|
|
|
0.27±0.06 |
||
|
Nanoparticles obtained by the hydrolysis of β-CN with trypsin |
Large |
235 |
95 |
0.40 |
|
400 |
93 |
0.23 |
||
|
437 |
207 |
0.47 |
||
|
370 |
172 |
0.46 |
||
|
314 |
62 |
0.20 |
||
|
408 |
170 |
0.42 |
||
|
|
|
0.36±0.12 |
||
|
Medium |
47 |
35 |
0.74 |
|
|
54 |
44 |
0.81 |
||
|
75 |
50 |
0.67 |
||
|
46 |
21 |
0.46 |
||
|
|
|
0.67±0.15 |
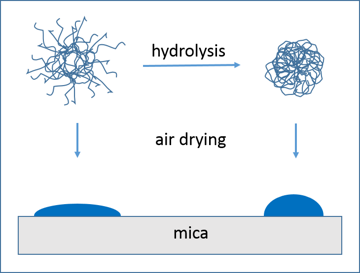
Figure 7. Change in H/D parameter of β-casein nanoparticles modified by hydrolysis with trypsin.
The formation of the denser axisymmetric particles is connected with the proteolytic action of trypsin: small micelles disappear with the formation of soluble peptides, small pieces are cleaved from the protruding parts of micelles and acquire more rounded shape. The rate of hydrolysis of Arg25-Ile26 peptide bond, which determines the elimination of the N-terminal fragment of β-CN, is several times higher than the rate of hydrolysis of the other trypsin-specific peptide bonds [7]. As a result of the loss of the charged N-terminal fragment, in which all the phosphoserine residues are concentrated, the rest of the β-CN polypeptide chain, including the hydrophobic C-terminal fragment, is more actively involved in the interchain interactions. It also determines the incorporation of hydrophobic peptide fragments and whole casein molecules, which were not previously a part of the micelles, into modified micelles. Thus, the action of trypsin can result in the formation of denser particles. The AFM data presented here are in accordance with this point.
Earlier we performed encapsulation of the biologically active compounds (BACs) into casein peptide nanoparticles in order to obtain water-soluble colloidal forms of BACs. β-Oxalylamino O-ethyl-N-aryl carbamates and N-ethyl-N'-arylureas, which represent a new class of chlorine-containing phytoactive compounds with potential growth-regulating activity of cereal crops, were used as BAC [10]. The highly sensitive coulometric method for chlorine determination was used to show that the nanoparticles contain the higher amount of chlorine-substituted alkyl carbamate than the original casein micelles [10].
The hydrolysis of β-CN by trypsin is a case in which the hydrolysis process is limited due to the large proportion of peptide bonds inside the nanoparticles that are hidden for the enzymatic attack (masked). The process of peptide bond demasking is important for the quantitative analysis of the kinetics of proteolysis for any enzyme–substrate pair [15]. The development of the quantitative method for analyzing nanoparticles from casein is also urgent for modeling the proteolysis and peptide bond demasking.
Experimental
General remarks
β-CN (C6905) from bovine milk and trypsin (T1426) from bovine pancreas of analytical grade treated with N-tosyl-l-phenylalanine chloromethyl ketone (TPCK) were purchased from Sigma–Aldrich. TPCK was used to inhibit contaminating chymotrypsin activity without affecting trypsin activity. Phosphate buffer solution was prepared with doubly distilled water and kept at 4 °C before use. Trypsin solutions were freshly prepared by diluting freeze-dried trypsin in phosphate buffer. All other reagents of analytical grade were obtained from commercial sources.
Proteolysis reaction and formation of nanoparticles
A typical proteolytic experiment leading to the formation of nanoparticles was as follows: the casein proteolysis with trypsin was carried out in 50 mM phosphate buffer at pH 7.9 under thermostatic conditions (37 °C) with magnetic stirring. Reconstitution of β-CN micelles was performed by solubilization of casein preparation in 50 mM phosphate buffer at 37 °C under slow stirring for at least 3 h. The hydrolysis of β-casein micelles at the concentration of 3.0 g/L in the volume of 10 mL was started by adding 10 μL of trypsin solution (1 g/L), and the reaction was terminated by adding the soybean trypsin inhibitor at the molar ratio of trypsin to inhibitor of 1:3 (w/w). To control the proteolysis, the degree of hydrolysis of peptide bonds (DH) in the samples taken from the reaction mixture was determined using OPA method [16]. This method is based on the reaction of o-phthalic aldehyde and 2-mercaptoethanol with amino groups formed during the hydrolysis of peptide bonds. OPA reagent was prepared by mixing 25 mL of 100 mM sodium tetrahydroborate, 2.5 mL of 20% SDS solution (w/w), 40 mg of o-phthalic aldehyde, and 100 μL of 2-mercaptoethanol. The final volume of OPA reagent was adjusted with distilled water to 50 mL. To each sample (40 μL) taken at the time of hydrolysis t, OPA reagent (2 mL) was added, and the resulting mixture was shaken and left at room temperature for 5 min. Then, the absorbance at 340 nm was determined. Based on the obtained values of absorption, the concentrations of amino nitrogen and the values of DH were determined at any time t. The proteolysis was stopped by adding the soybean inhibitor when the desired DH value was achieved.
Atomic force microscopy
The analysis of the nanoparticles obtained by atomic force microscopy was performed using a FemtoScan microscope. The samples, taken from the reaction mixture, were diluted 10 times with water (Milli-Q). Immediately after this, the analyzed sample (2.5 μL) was applied to the freshly cleaved surface of mica and dried in air. The samples were examined under ambient conditions in tapping mode. The experiments were carried out using FS_SS_HR cantilevers (ScanSens, series ETALON) with resonant frequencies of about 230 and 380 kHz and probe radius of about 10 nm. The images with the sizes of 4×4 µm2 and 1×1 µm2 were scanned to measure the sizes of the large and medium peptide particles, respectively. The AFM images were processed and analyzed using FemtoScan Online software [17]. To measure the diameter and height of the particles, the sections of the surface were constructed, passing through the point of maximum particle height and directed at the angle of 45° to the fast scanning axis.
Conclusions
The changes in the morphology of β-casein nanoparticles after tryptic hydrolysis were elucidated using atomic force microscopy. The resulting nanoparticles were found to be approximately two times denser than the initial casein micelles. These results are useful for analyzing the mechanism of proteolysis of heterogeneous protein substrates and are also important for the design of new biocompatible nontoxic systems for transporting biologically active compounds.
Acknowledgements
This work was supported by the Ministry of Science and Higher Education of the Russian Federation.
References
- L. Ya. Zakharova, R. R. Kashapov, T. N. Pashirova, A. B. Mirgorodskaya, O. G. Sinyashin, Mendeleev Commun., 2016, 26, 457–468. DOI: 10.1016/j.mencom.2016.11.001
- D. G. Dalgleish, Soft Matter, 2011, 7, 2265–2272. DOI: 10.1039/C0SM00806K
- E. Semo, E. Kesselman, D. Danino, Y. D. Livney, Food Hydrocolloids, 2007, 21, 936–942. DOI: 10.1016/j.foodhyd.2006.09.006
- Y. D. Livney, Curr. Opin. Colloid Interface Sci., 2010, 15, 73–83. DOI: 10.1016/j.cocis.2009.11.002
- C. Holt, L. Sawyer, J. Chem. Soc., Faraday Trans., 1993, 89, 2683–2692. DOI: 10.1039/FT9938902683
- D. S. Horne, in: Milk Proteins, 2nd ed., H. Singh, M. Boland, A. Thompson (Eds.), Acad. Press, 2014, 169–200. DOI: 10.1016/B978-0-12-405171-3.00006-4
- M. M. Vorob'ev, M. Dalgalarrondo, J.-M. Chobert, T. Haertlé, Biopolymers, 2000, 54, 355–364. DOI: 10.1002/1097-0282(20001015)54:5<355::AID-BIP60>3.0.CO;2-H
- M. M. Vorob'ev, V. Vogel, W. Mäntele, Int. Dairy J., 2013, 30, 33–38. DOI: 10.1016/j.idairyj.2012.12.002
- М. М. Vorob'ev, K. Strauss, V. Vogel, W. Mäntele, Food Biophys., 2015, 10, 309–315. DOI: 10.1007/s11483-015-9391-6
- M. M. Vorob'ev, V. S. Khomenkova, O. V. Sinitsyna, О. А. Levinskaya, D. Kh. Kitaeva, A. V. Kalistratova, M. S. Oshchepkov, L. V. Kovalenko, K. A. Kochetkov, Russ. Chem. Bull., 2018, 67, 1508–1512. DOI: 10.1007/s11172-018-2248-7
- G. Binnig, C. F. Quate, Ch. Gerber, Phys. Rev. Lett., 1986, 56, 930–933. DOI: 10.1103/PhysRevLett.56.930
- A. P. Gunning, V. J. Morris, Food Hydrocolloids, 2018, 78, 62–76. DOI: 10.1016/j.foodhyd.2017.05.017
- C. Shi, Y. He, M. Ding, Y. Wang, J. Zhong, Trends Food Sci. Technol., 2018, 87, 14–25. DOI: 10.1016/j.tifs.2018.11.027
- M. Ouanezar, F. Guyomarc'h, A. Bouchoux, Langmuir, 2012, 28, 4915–4919. DOI: 10.1021/la3001448
- M. M. Vorob'ev, J. Mol. Catal. B: Enzym., 2009, 58, 146–152. DOI: 10.1016/j.molcatb.2008.12.007
- F. C. Church, D. H. Porter, G. L. Catignani, H. E. Swaisgood, Anal. Biochem., 1985, 146, 343–348. DOI: 10.1016/0003-2697(85)90549-4
- I. V. Yaminsky, A. I. Akhmetova, G. B. Meshkov, Nanoindustry, 2018, 11 (6), 414–416. DOI: 10.22184/1993-8578.2018.11.6.414.416





